Huimin Li
Learning with Geometric Priors in U-Net Variants for Polyp Segmentation
Jan 24, 2026Abstract:Accurate and robust polyp segmentation is essential for early colorectal cancer detection and for computer-aided diagnosis. While convolutional neural network-, Transformer-, and Mamba-based U-Net variants have achieved strong performance, they still struggle to capture geometric and structural cues, especially in low-contrast or cluttered colonoscopy scenes. To address this challenge, we propose a novel Geometric Prior-guided Module (GPM) that injects explicit geometric priors into U-Net-based architectures for polyp segmentation. Specifically, we fine-tune the Visual Geometry Grounded Transformer (VGGT) on a simulated ColonDepth dataset to estimate depth maps of polyp images tailored to the endoscopic domain. These depth maps are then processed by GPM to encode geometric priors into the encoder's feature maps, where they are further refined using spatial and channel attention mechanisms that emphasize both local spatial and global channel information. GPM is plug-and-play and can be seamlessly integrated into diverse U-Net variants. Extensive experiments on five public polyp segmentation datasets demonstrate consistent gains over three strong baselines. Code and the generated depth maps are available at: https://github.com/fvazqu/GPM-PolypSeg
Topo-VM-UNetV2: Encoding Topology into Vision Mamba UNet for Polyp Segmentation
May 09, 2025



Abstract:Convolutional neural network (CNN) and Transformer-based architectures are two dominant deep learning models for polyp segmentation. However, CNNs have limited capability for modeling long-range dependencies, while Transformers incur quadratic computational complexity. Recently, State Space Models such as Mamba have been recognized as a promising approach for polyp segmentation because they not only model long-range interactions effectively but also maintain linear computational complexity. However, Mamba-based architectures still struggle to capture topological features (e.g., connected components, loops, voids), leading to inaccurate boundary delineation and polyp segmentation. To address these limitations, we propose a new approach called Topo-VM-UNetV2, which encodes topological features into the Mamba-based state-of-the-art polyp segmentation model, VM-UNetV2. Our method consists of two stages: Stage 1: VM-UNetV2 is used to generate probability maps (PMs) for the training and test images, which are then used to compute topology attention maps. Specifically, we first compute persistence diagrams of the PMs, then we generate persistence score maps by assigning persistence values (i.e., the difference between death and birth times) of each topological feature to its birth location, finally we transform persistence scores into attention weights using the sigmoid function. Stage 2: These topology attention maps are integrated into the semantics and detail infusion (SDI) module of VM-UNetV2 to form a topology-guided semantics and detail infusion (Topo-SDI) module for enhancing the segmentation results. Extensive experiments on five public polyp segmentation datasets demonstrate the effectiveness of our proposed method. The code will be made publicly available.
Adapting a Segmentation Foundation Model for Medical Image Classification
May 09, 2025



Abstract:Recent advancements in foundation models, such as the Segment Anything Model (SAM), have shown strong performance in various vision tasks, particularly image segmentation, due to their impressive zero-shot segmentation capabilities. However, effectively adapting such models for medical image classification is still a less explored topic. In this paper, we introduce a new framework to adapt SAM for medical image classification. First, we utilize the SAM image encoder as a feature extractor to capture segmentation-based features that convey important spatial and contextual details of the image, while freezing its weights to avoid unnecessary overhead during training. Next, we propose a novel Spatially Localized Channel Attention (SLCA) mechanism to compute spatially localized attention weights for the feature maps. The features extracted from SAM's image encoder are processed through SLCA to compute attention weights, which are then integrated into deep learning classification models to enhance their focus on spatially relevant or meaningful regions of the image, thus improving classification performance. Experimental results on three public medical image classification datasets demonstrate the effectiveness and data-efficiency of our approach.
Self Pre-training with Topology- and Spatiality-aware Masked Autoencoders for 3D Medical Image Segmentation
Jun 15, 2024



Abstract:Masked Autoencoders (MAEs) have been shown to be effective in pre-training Vision Transformers (ViTs) for natural and medical image analysis problems. By reconstructing missing pixel/voxel information in visible patches, a ViT encoder can aggregate contextual information for downstream tasks. But, existing MAE pre-training methods, which were specifically developed with the ViT architecture, lack the ability to capture geometric shape and spatial information, which is critical for medical image segmentation tasks. In this paper, we propose a novel extension of known MAEs for self pre-training (i.e., models pre-trained on the same target dataset) for 3D medical image segmentation. (1) We propose a new topological loss to preserve geometric shape information by computing topological signatures of both the input and reconstructed volumes, learning geometric shape information. (2) We introduce a pre-text task that predicts the positions of the centers and eight corners of 3D crops, enabling the MAE to aggregate spatial information. (3) We extend the MAE pre-training strategy to a hybrid state-of-the-art (SOTA) medical image segmentation architecture and co-pretrain it alongside the ViT. (4) We develop a fine-tuned model for downstream segmentation tasks by complementing the pre-trained ViT encoder with our pre-trained SOTA model. Extensive experiments on five public 3D segmentation datasets show the effectiveness of our new approach.
DNANet: De-Normalized Attention Based Multi-Resolution Network for Human Pose Estimation
Oct 21, 2019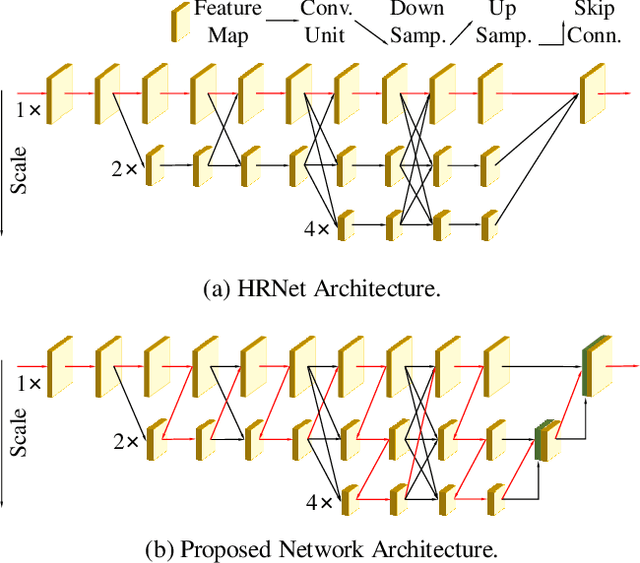
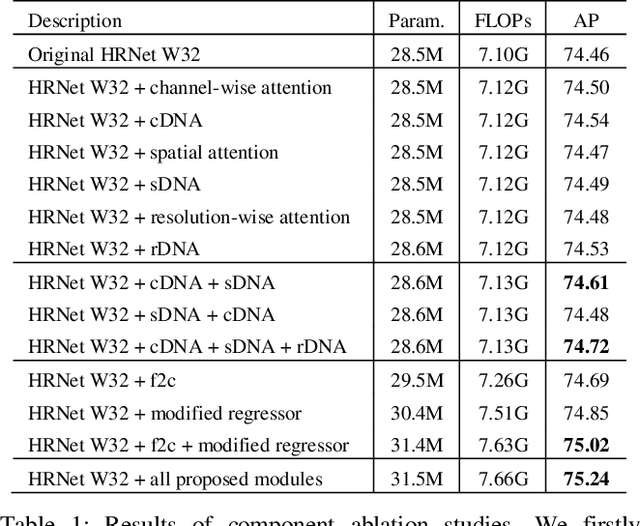
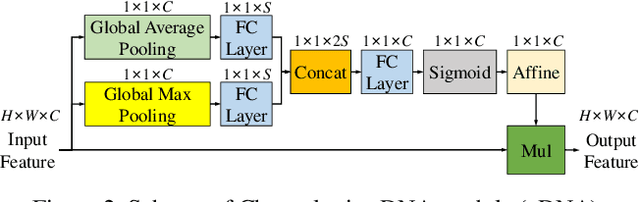
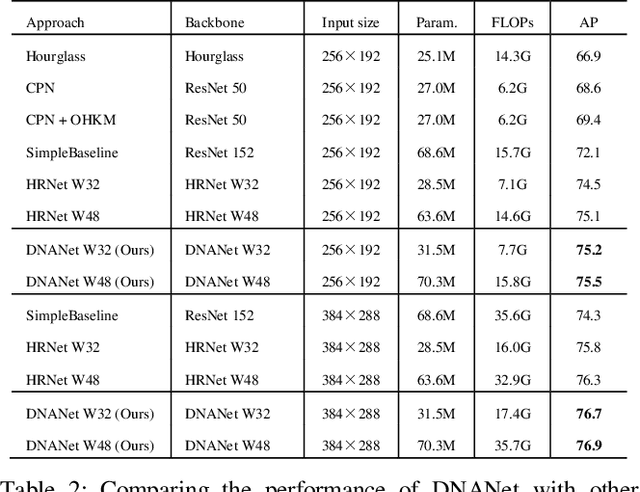
Abstract:Recently, multi-resolution networks (such as Hourglass, CPN, HRNet, etc.) have achieved significant performance on the task of human pose estimation by combining features from various resolutions. In this paper, we propose a novel type of attention module, namely De-Normalized Attention (DNA) to deal with the feature attenuations of conventional attention modules. Our method extends the original HRNet with spatial, channel-wise and resolution-wise DNAs, which aims at evaluating the importance of features from different locations, channels and resolutions to enhance the network capability for feature representation. We also propose to add fine-to-coarse connections across high-to-low resolutions in-side each layer of HRNet to increase the maximum depth of network topology. In addition, we propose to modify the keypoint regressor at the end of HRNet for accurate keypoint heatmap prediction. The effectiveness of our proposed network is demonstrated on COCO keypoint detection dataset, achieving state-of-the-art performance at 77.6 AP score on COCO val2017 dataset without using extra keypoint training data. Our paper will be accompanied with publicly available codes at GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge